Fixed concentration boundary
import pyrid as prd
file_path='Files/'
fig_path = 'Figures/'
file_name='Fixed_Concentration'
nsteps = 1e5
stride = int(nsteps/1000)
obs_stride = int(nsteps/1000)
box_lengths = [250.0,250.0,350.0]
Temp=293.15
eta=1e-21
dt = 10.0
Simulation = prd.Simulation(box_lengths = box_lengths,
dt = dt,
Temp = Temp,
eta = eta,
stride = stride,
write_trajectory = True,
file_path = file_path,
file_name = file_name,
fig_path = fig_path,
boundary_condition = 'fixed concentration',
nsteps = nsteps,
seed = 0,
length_unit = 'nanometer',
time_unit = 'ns')
Simulation.register_particle_type('Core_0', 2.5)
Simulation.register_particle_type('Core_1', 2.5)
Simulation.register_particle_type('Core_2', 2.5)
vertices, triangles, Compartments = prd.load_compartments('Compartments/Synapse.obj')
Simulation.set_compartments(Compartments, triangles, vertices, mesh_scale = 1e3/2)
prd.plot.plot_compartments(Simulation, save_fig = False, show = True)
A_pos = np.array([[0.0,0.0,0.0]])
A_types = np.array(['Core_0'], dtype = np.dtype('U20'))
B_pos = np.array([[0.0,0.0,0.0]])
B_types = np.array(['Core_1'], dtype = np.dtype('U20'))
C_pos = np.array([[0.0,0.0,0.0]])
C_types = np.array(['Core_2'], dtype = np.dtype('U20'))
Simulation.register_molecule_type('A', A_pos, A_types, collision_type = 1)
D_tt, D_rr = prd.diffusion_tensor(Simulation, 'A')
Simulation.set_diffusion_tensor('A', D_tt, D_rr)
Simulation.register_molecule_type('B', B_pos, B_types, collision_type = 1)
D_tt, D_rr = prd.diffusion_tensor(Simulation, 'B')
Simulation.set_diffusion_tensor('B', D_tt, D_rr)
Simulation.register_molecule_type('C', C_pos, C_types, collision_type = 1)
D_tt, D_rr = prd.diffusion_tensor(Simulation, 'C')
Simulation.set_diffusion_tensor('C', D_tt, D_rr)
concentration = 100/Simulation.System.volume
Simulation.fixed_concentration_at_boundary('A', concentration, 'Box', 'Volume')
concentration = 100/Simulation.System.Compartments[1].volume
Simulation.fixed_concentration_at_boundary('B', concentration, 'Postsynapse', 'Volume')
concentration = 100/Simulation.System.Compartments[1].area
Simulation.fixed_concentration_at_boundary('C', concentration, 'Postsynapse', 'Surface')
Simulation.observe('Number', molecules = ['A', 'B', 'C'], obs_stride = obs_stride)
Simulation.run(progress_stride = 1000, out_linebreak = False)
Simulation.print_timer()
Evaluation = prd.Evaluation()
Evaluation.load_file(file_name)
import matplotlib.pyplot as plt
Evaluation.plot_observable('Number', molecules = ['A', 'B', 'C'], save_fig = True)
plt.axhline(1000, color = 'k', linestyle = '--', linewidth = 1, zorder = 0)
plt.savefig('Figures//Fixed_Concentration_Number.png', bbox_inches="tight", dpi = 300)
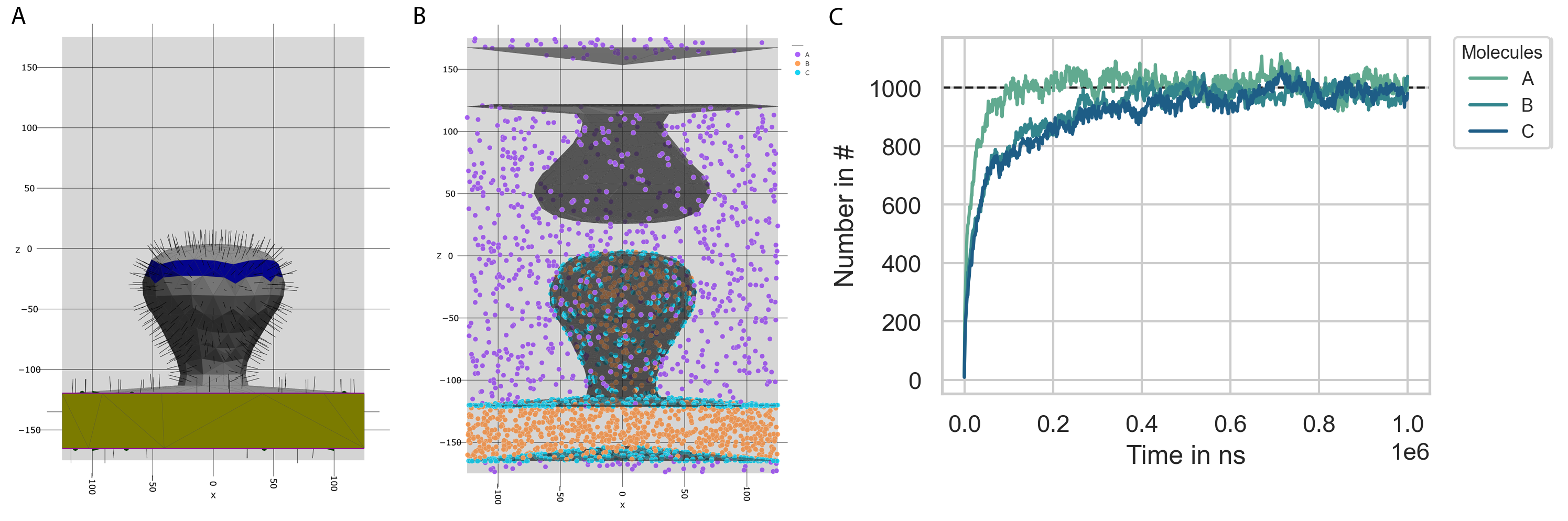
Fig. 10 The molecule concentration inside the simulation box approaches the outside molecule concentration (C).