Large scale toy-model
import pyrid as prd
file_path='Files/'
fig_path = 'Figures/'
file_name='Large_Toy_Model'
nsteps = 1e4
stride = int(nsteps/100)
obs_stride = int(nsteps/100)
use_checkpoint = False # True #
checkp = False
box_lengths = [250.0,250.0,350.0]
Temp=293.15
eta=1e-21
dt = 0.1
Simulation = prd.Simulation(box_lengths = box_lengths,
dt = dt,
Temp = Temp,
eta = eta,
stride = stride,
write_trajectory = True,
file_path = file_path,
file_name = file_name,
fig_path = fig_path,
boundary_condition = 'periodic',
nsteps = nsteps,
seed = 1000,
wall_force = 100.0,
length_unit = 'nanometer',
time_unit = 'ns')
Simulation.add_checkpos(int(nsteps/10), "checkpos/", 1) # stride, directory, max_saves
Simulation.register_particle_type('Core_1', 1.5) # (Name, Radius)
Simulation.register_particle_type('Core_1a', 1.5)
Simulation.register_particle_type('Patch_1', 0.0)
Simulation.register_particle_type('Patch_2', 0.0)
Simulation.register_particle_type('Core_2', 2.5)
Simulation.register_particle_type('Core_2a', 2.5)
Simulation.register_particle_type('Core_3', 1.0)
Simulation.register_particle_type('Core_4', 0.5)
Simulation.register_particle_type('Core_5', 0.75)
Simulation.register_particle_type('Core_6', 0.75)
Simulation.register_particle_type('Patch_3', 0.0)
A_pos = [[0.0,0.0,1.5], [0.0,0.0,-1.5], [-1.5,0.0,1.5], [1.5,0.0,-1.5]]
A_types = ['Core_1','Core_1','Patch_1','Patch_2']
B_pos = prd.distribute_surf.evenly_on_sphere(4,2.5)
B_types = ['Core_2','Patch_1','Patch_1', 'Patch_1', 'Patch_1']
B2_pos = [[0.0,0.0,1.5], [0.0,0.0,-2.5], [-1.5,0.0,1.5], [1.5,0.0,1.5], [2.5,0.0,-2.5]]
B2_types = ['Core_1a','Core_2a','Patch_1','Patch_1','Patch_2']
C_pos = [[0.0,0.0,0.0]]
C_types = ['Core_3']
D_pos = [[0.0,0.0,0.0]]
D_types = ['Core_4']
E_pos =[[0.0,0.0,0.0]]
E_types =['Core_5']
F_pos =[[0.0,0.0,0.0]]
F_types = ['Core_6']
Simulation.register_molecule_type('A', A_pos, A_types)
Simulation.register_molecule_type('B', B_pos, B_types)
Simulation.register_molecule_type('B2', B2_pos, B2_types)
Simulation.register_molecule_type('C', C_pos, C_types, 1)
Simulation.register_molecule_type('D', D_pos, D_types, 1)
Simulation.register_molecule_type('E', E_pos, E_types, 1)
Simulation.register_molecule_type('F', F_pos, F_types, 1)
D_tt, D_rr, D_tr, D_rt, rOD_A, rCoM_A = prd.diffusion_tensor(Simulation, 'A', return_CoD = True, return_coupling = True, return_CoM = True)
Simulation.set_diffusion_tensor('A', D_tt, D_rr)
D_tt, D_rr, D_tr, D_rt, rOD_B, rCoM_B = prd.diffusion_tensor(Simulation, 'B', True, True, True)
Simulation.set_diffusion_tensor('B', D_tt, D_rr)
D_tt, D_rr, D_tr, D_rt, rOD_B2, rCoM_B2 = prd.diffusion_tensor(Simulation, 'B2', True, True, True)
Simulation.set_diffusion_tensor('B2', D_tt, D_rr)
D_tt, D_rr, D_tr, D_rt, rOD_C, rCoM_C = prd.diffusion_tensor(Simulation, 'C', True, True, True)
Simulation.set_diffusion_tensor('C', D_tt, D_rr)
D_tt, D_rr, D_tr, D_rt, rOD_D, rCoM_D = prd.diffusion_tensor(Simulation, 'D', True, True, True)
Simulation.set_diffusion_tensor('D', D_tt, D_rr)
D_tt, D_rr, D_tr, D_rt, rOD_E, rCoM_E = prd.diffusion_tensor(Simulation, 'E', True, True, True)
Simulation.set_diffusion_tensor('E', D_tt, D_rr)
D_tt, D_rr, D_tr, D_rt, rOD_F, rCoM_F = prd.diffusion_tensor(Simulation, 'F', True, True, True)
Simulation.set_diffusion_tensor('F', D_tt, D_rr)
prd.plot.plot_mobility_matrix('A', Simulation, save_fig = True, show = True)
prd.plot.plot_mobility_matrix('B', Simulation, save_fig = True, show = True)
prd.plot.plot_mobility_matrix('B2', Simulation, save_fig = True, show = True)
prd.plot.plot_mobility_matrix('C', Simulation, save_fig = True, show = True)
#-----------------------------------------------------
# Add Global Pair Interactions
#-----------------------------------------------------
k=100.0 #kJ/(avogadro*nm^2)
Simulation.add_interaction('harmonic_repulsion', 'Core_1', 'Core_1', {'k':k}, bond = False)
Simulation.add_interaction('harmonic_repulsion', 'Core_1', 'Core_2', {'k':k}, bond = False)
Simulation.add_interaction('harmonic_repulsion', 'Core_2', 'Core_2', {'k':k}, bond = False)
#-----------------------------------------------------
# Add Pair Binding Reaction
#-----------------------------------------------------
k=100.0
h=50.0
d=0.0
rc = 2.0
Simulation.add_bp_reaction('bind', ['Patch_1', 'Patch_3'], ['Patch_1', 'Patch_3'], 10.0, 2.0, 'harmonic_attraction', {'k':k, 'h':h , 'rc':rc})
Simulation.add_bp_reaction('bind', ['Patch_1', 'Patch_2'], ['Patch_1', 'Patch_2'], 10.0, 2.0, 'harmonic_attraction', {'k':k, 'h':h , 'rc':rc})
Simulation.add_bp_reaction('bind', ['Patch_1', 'Patch_1'], ['Patch_1', 'Patch_1'], 10.0, 2.0, 'harmonic_attraction', {'k':k, 'h':h , 'rc':rc})
Simulation.add_bp_reaction('bind', ['Patch_2', 'Patch_3'], ['Patch_2', 'Patch_3'], 10.0, 2.0, 'harmonic_attraction', {'k':k, 'h':h , 'rc':rc})
Simulation.add_bp_reaction('bind', ['Patch_2', 'Patch_2'], ['Patch_2', 'Patch_2'], 10.0, 2.0, 'harmonic_attraction', {'k':k, 'h':h , 'rc':rc})
Simulation.add_bp_reaction('bind', ['Patch_3', 'Patch_3'], ['Patch_3', 'Patch_3'], 10.0, 2.0, 'harmonic_attraction', {'k':k, 'h':h , 'rc':rc})
prd.plot.plot_potential(Simulation, [(prd.potentials.harmonic_repulsion, [3.0,k]), (prd.potentials.piecewise_harmonic, [2.0,k,h,d])], yU_limits = [-60,300], yF_limits = [-60 ,300 ], r_limits = [0,4], save_fig = True)
#%%
#-----------------------------------------------------
# Add Particle Conversion Reaction
#-----------------------------------------------------
Simulation.add_up_reaction('conversion', 'Patch_1', 0.01, ['Patch_2'])
#-----------------------------------------------------
# Add Particle Enzymatic Reaction
#-----------------------------------------------------
Simulation.add_bp_reaction('enzymatic', ['Patch_2', 'Core_6'], ['Patch_3', 'Core_6'], 1.0, 3.0)
#-----------------------------------------------------
# Add Molecule Production Reaction
#-----------------------------------------------------
Simulation.add_um_reaction('production', 'A', 0.01, ['B2']+['D']*5+['C']*5+['E']*10+['B']*2, [1]+[0]*5+[0]*5+[1]*10+[1]*2, [1]+[1]*5+[-1]*5+[1]*10+[1]*2, 10.0)
#-----------------------------------------------------
# Add Molecule Fusion Reaction
#-----------------------------------------------------
Simulation.add_bm_reaction('fusion', ['B', 'B'], ['A'], [['Core_2', 'Core_2']], [0.1], [5.0])
Simulation.add_bm_reaction('fusion', ['B', 'B'], ['C'], [['Core_2', 'Core_2']], [0.1], [5.0])
Simulation.add_bm_reaction('fusion', ['B', 'B'], ['C'], [['Core_2', 'Core_2']], [0.05], [5.0])
Simulation.add_bm_reaction('fusion', ['A', 'B'], ['B2'], [['Core_1', 'Core_2']], [0.01], [5.0])
Simulation.add_bm_reaction('fusion', ['A', 'B2'], ['C'], [['Core_1', 'Core_2a']], [0.01], [5.0])
Simulation.add_bm_reaction('fusion', ['A', 'B2'], ['C'], [['Core_1', 'Core_1a']], [0.05], [5.0])
#-----------------------------------------------------
# Add Molecule Enzymatic Reaction
#-----------------------------------------------------
Simulation.add_bm_reaction('enzymatic_mol', ['C', 'E'], ['F', 'E'], [['Core_3', 'Core_5']], [0.1], [3.0])
#%%
Evaluation = prd.Evaluation()
Evaluation.plot_reactions_graph(Simulation, graph_type = 'Bimolecular')
Evaluation.plot_reactions_graph(Simulation, graph_type = 'Interactions')
Evaluation.plot_reactions_graph(Simulation, graph_type = 'Unimolecular')
vertices, triangles, Compartments = prd.load_compartments('Compartments/Synapse.obj')
Simulation.set_compartments(Compartments, triangles, vertices, mesh_scale = 1e3/2)
prd.plot.plot_compartments(Simulation, save_fig = True, show = False)
#-----------------------------------------------------
# Add a release event
#-----------------------------------------------------
Simulation.add_release_site('Volume', 1000, 0, Number = [300,50], Types = ['C', 'D'], origin = [0.0, 0.0, 24.5], jitter = 1.0)
Simulation.add_release_site('Surface', 2000, 1, Number = [500], Types = ['D'], triangle_id = Simulation.System.Compartments[1].triangle_ids[50], jitter = 5.0)
if checkp == False:
pos, mol_type_idx, quaternion = Simulation.distribute('PDS uniform', 'Volume', 0, ['B'], [2000], multiplier = 50)
Simulation.add_molecules('Volume',0, pos, quaternion, mol_type_idx)
# prd.plot.plot_sphere_packing(0, Simulation, pos, mol_type_idx)
# Postysnapse:
pos, mol_type_idx, quaternion = Simulation.distribute('PDS uniform', 'Volume', 1, ['B'], [500], multiplier = 50)
Simulation.add_molecules('Volume',1, pos, quaternion, mol_type_idx)
# prd.plot.plot_sphere_packing(1, Simulation, pos, mol_type_idx)
pos, mol_type_idx, quaternion, face_ids = Simulation.distribute('MC', 'Surface', 1, ['A', 'B'], [200,200])
Simulation.add_molecules('Surface',1, pos, quaternion, mol_type_idx, face_ids)
# prd.plot.plot_sphere_packing(1, Simulation, pos, mol_type_idx)
# Presynapse:
pos, mol_type_idx, quaternion = Simulation.distribute('PDS uniform', 'Volume', 2, ['B'], [500], multiplier = 50)
Simulation.add_molecules('Volume',2, pos, quaternion, mol_type_idx)
# prd.plot.plot_sphere_packing(1, Simulation, pos, mol_type_idx)
pos, mol_type_idx, quaternion, face_ids = Simulation.distribute('PDS', 'Surface', 2, ['A', 'B'], [200,200])
Simulation.add_molecules('Surface',2, pos, quaternion, mol_type_idx, face_ids)
else:
Simulation.load_checkpoint('Large_Toy_Model', 0)
prd.plot.plot_scene(Simulation, save_fig = True, show = False)
Simulation.observe_rdf(rdf_pairs = [['A','A'],['A','B'],['A','C']], rdf_bins = [100,100,100], rdf_cutoff = [20.0,20.0,20.0], stride = obs_stride)
Simulation.observe('Force', molecules = ['A', 'B', 'C'], obs_stride = obs_stride, binned = True)
Simulation.observe('Torque', molecules = ['A', 'B', 'C'], obs_stride = obs_stride)
Simulation.observe('Energy', obs_stride = obs_stride, binned = False)
Simulation.observe('Volume', obs_stride = obs_stride, binned = False)
Simulation.observe('Pressure', molecules = ['A'], obs_stride = obs_stride, binned = False)
Simulation.observe('Virial', molecules = ['A'], obs_stride = obs_stride, binned = False)
Simulation.observe('Virial Tensor', molecules = ['A'], obs_stride = obs_stride, binned = False)
Simulation.observe('Number', molecules = ['A', 'B', 'C', 'B2', 'D', 'E', 'F'], obs_stride = obs_stride, binned = False)
Simulation.observe('Reactions', obs_stride = obs_stride, binned = True)
Simulation.observe('Bonds', obs_stride = obs_stride)
Simulation.observe('Orientation', molecules = ['A'], obs_stride = obs_stride)
Simulation.observe('Position', molecules = ['A'], obs_stride = obs_stride)
Simulation.run(progress_stride = 1000, out_linebreak = False)
Simulation.print_timer()
prd.plot.plot_concentration_profile(Simulation, axis = 0, save_fig = True)
#%%
Evaluation.load_file(file_name)
Evaluation.read_observable('Number')
Evaluation.read_observable('Bonds')
Evaluation.read_observable('Reactions', Reaction_Type = 'bind')
Evaluation.read_observable('Reactions', Reaction_Type = 'fusion')
#%%
Evaluation.plot_observable('Reactions', Reaction_Type = 'bind', educt = 'Patch_1+Patch_3', save_fig = True)
Evaluation.plot_observable('Reactions', Reaction_Type = 'enzymatic', educt = 'Patch_2+Core_6', save_fig = True)
Evaluation.plot_observable('Reactions', Reaction_Type = 'fusion', educt = 'A+B2', particle_educt = 'Core_1+Core_2a', save_fig = True)
Evaluation.plot_observable('Bonds', bond_pairs = 'All', save_fig = True)
Evaluation.plot_observable('Number', molecules = ['A', 'B', 'C'], save_fig = True)
Evaluation.plot_observable('Energy', save_fig = True)
Evaluation.plot_observable('Pressure', save_fig = True)
Evaluation.plot_observable('Virial', save_fig = True)
#%%
Evaluation.plot_observable('Force', molecules = ['B'], step = 10, save_fig = True)
Evaluation.plot_observable('Torque', molecules = ['B'], step = 10, save_fig = True)
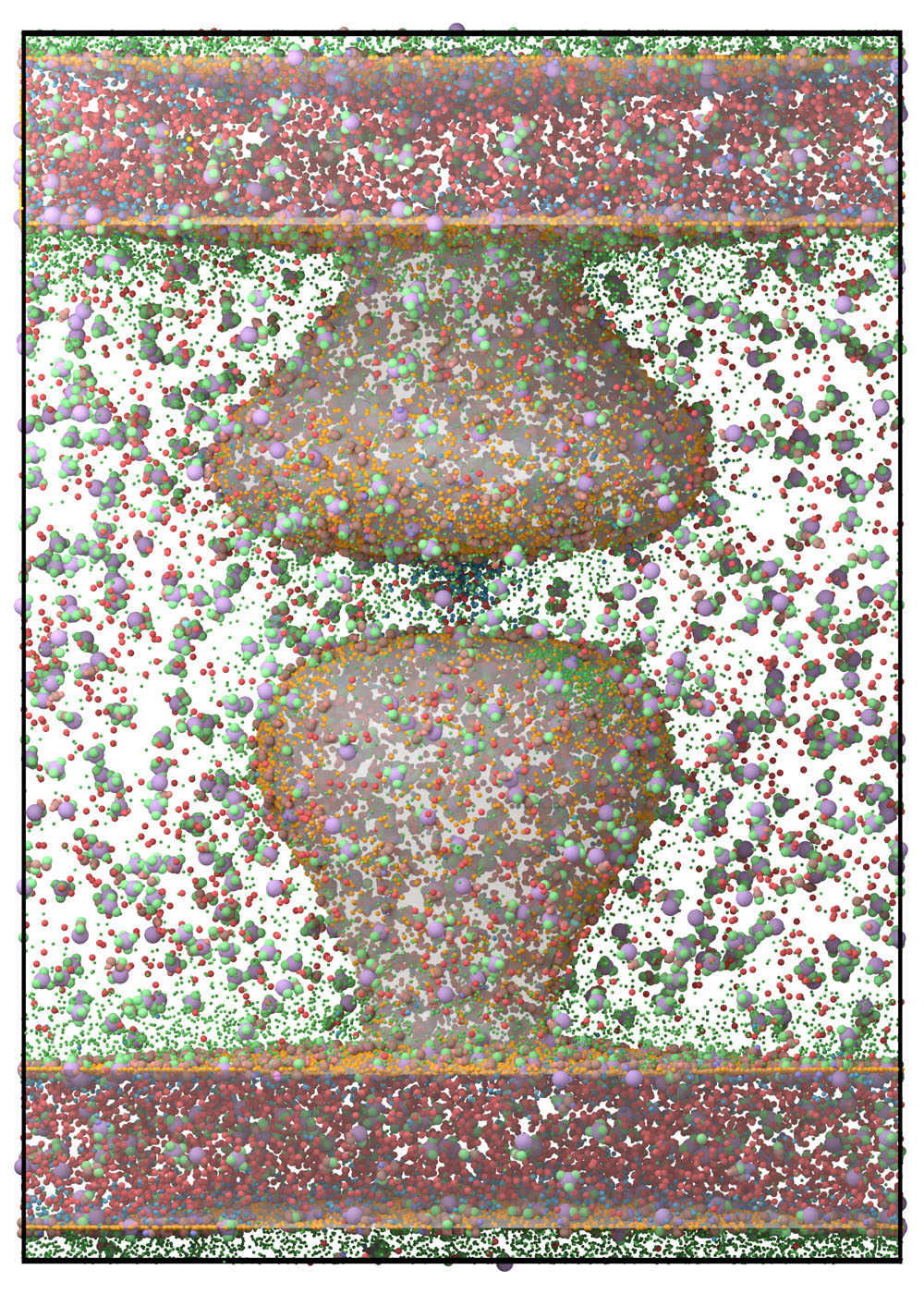
Fig. 9 Render.